Protein Dynamics and Substrate Protonation State Mediate the Catalytic Action of Trans-4-Hydroxy-L-Proline Dehydratase
Abstract
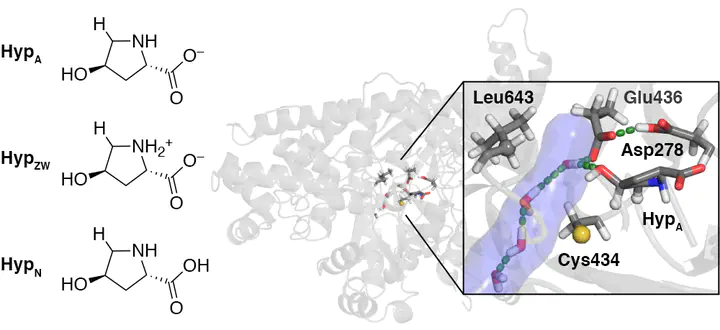
The enzyme trans-4-Hydroxy-L-proline (Hyp) dehydratase (HypD) is among the most abundant glycyl radical enzymes (GREs) in the healthy human gut microbiome and is considered a promising antibiotic target for the prominent antibiotic-resistant pathogen Clostridium difficile. Although an enzymatic mechanism has been proposed, the role of the greater HypD protein environment in mediating radical reactivity is not well understood. To fill this gap in understanding, we investigate HypD across multiple time- and length- scales using electronic structure modeling and classical molecular dynamics. We observe that the Hyp substrate protonation state significantly alters both its enzyme-free reactivity and its dynamics within the enzyme active site. Accurate coupled cluster modeling suggests the deprotonated form of Hyp to be the most reactive protonation state for C5–Hpro-S activation. In the protein environment, hydrophobic interactions modulate the positioning of Cys434 radical to enhance the reactivity of C5–Hpro-S abstraction. Long-time dynamics reveal that changing Hyp protonation states triggers the switching of a Leu643-gated water tunnel, a functional feature that has not yet been observed for members of the GRE superfamily.