Spectroscopically Guided Simulations Reveal Distinct Strategies for Positioning Substrates to Achieve Selectivity in Nonheme Fe(II)/α-Ketoglutarate-Dependent Halogenases
Abstract
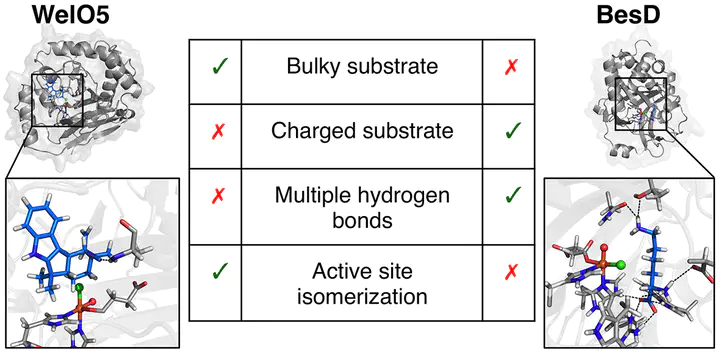
Nonheme iron halogenases, such as SyrB2, WelO5, and BesD, halogenate unactivated carbon atoms of diverse substrates at ambient conditions with exquisite selectivity seldom matched by nonbiological catalysts. Using experimentally guided molecular dynamics (MD) simulations augmented with multiscale (i.e., quantum mechanics/molecular mechanics) simulations of substrate-bound complexes of BesD and WelO5, we investigate substrate/active-site dynamics that enable selective halogenation. Our simulations reveal that active-site configurational isomerization is necessary in WelO5 to attain a substrate/active-site geometry consistent with its observed chemo- and regioselectivity. Conversely, a slight reorientation of the substrate from its crystal structure position is sufficient to enable regioselective chlorination in BesD without the need to invoke active-site isomerization. We observe very different patterns of substrate–protein interactions for these two enzymes, and we relate the nature of these interactions to the distinct substrates. For BesD, we resolve the uncertainty around the mechanistic relevance of Asn219. Our simulations reveal that the optimum substrate/active-site geometry also outweighs the interactions between the metal-oxo and the protein environment in facilitating the required chemoselectivity in halogenases. Our work highlights how different substrate-dependent strategies are used to accomplish selectivity-promoting proximity in halogenases.