molSimplify Tutorial 13: molscontrol -- an intelligent job control system to manage your DFT geometry optimizations for inorganic discovery.
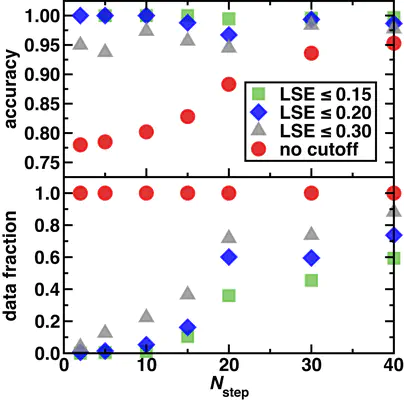
With the static classifier, we achieve great performance on job status predictions of out-of-sample data points, capturing their failure before doing the simulation thus saving lots of computational resources (Tutorial 12). When facing the out-of-distribution data that are dissimilar to the training data, which imitates more a chemistry discovery, the predictability of the static classifier is inevitably worsened as it is challenged by complexes of different composition. This asks for the existence of a model that can generalize better to fulfill the needs of chemical discovery. We built a dynamic classifier, which is essentially a convolutional neural network that takes the real-time DFT outputs step by step and makes the prediction on the final job status. Since the dynamic classifier uses the geometric and electronic structure of the DFT level as inputs, it is extremely transferable in terms of different chemical space and systematically improves with the increasing number of steps in geometry optimizations. Combined with the latent space entropy as a measure of model confidence, we built a job control system that can offer an “effective” two-fold acceleration for your calculation by killing simulations with a low expected success rate. More details please refer to our paper published at JCTC (https://pubs.acs.org/doi/10.1021/acs.jctc.9b00057).
This job control system, named molscontrol, is implemented in our open-source package molSimplify (https://github.com/hjkgrp/molSimplify). For molSimplify users, please either (re)run python setup.py install if you installed molSimplify from the source or download our most recent conda version of molSimplify to activate molscontrol.
molscontrol is a job control system so it needs to be run with a quantum chemistry process in parallel. An example bash script to do geometry optimization with Terachem is:
module load anaconda
source activate mols_keras
module load terachem/tip
terachem terachem_input > dft.out &
PID_KILL=$!
molscontrol $PID_KILL > control.out &
wait
where molscontrol takes in the PID of the terachem simulation in and monitors this process on-the-fly. Prediction probability and latent space entropy at each step are recorded in the log file. Besides the PID of your quantum chemistry calculation, molscontrol requires a configure file in your current working directory where the jobscript is submitted. Examples of configure.json can be found in the molSimplify/molscontrol/tests folder and each keyword is explained in detail at molSimplify/molscontrol/dynamic\_classifier.py.
molscontrol has the capacity of interfacing with other quantum chemistry packages by varying and customizing the “mode”. Currently two modes are implemented, “terachem” and “geo”. In “terachem” mode, the dynamic classifier utilizes outputs of gradients, Mulliken charges, bond order matrix and geometry (all in terachem printout format) at each step to make predictions on the final job label. For non-Terachem users, a “geo” mode is available where only the optimization trajectory is required as the input for the dynamic classifier, which should be readily obtained in most of the quantum chemistry packages. In addition, users are also welcomed to implement your own models based on the outputs of your quantum chemistry package of choice by filling out the “custom” dictionary in molSimplify/molscontrol/dynamic\_classifier.py.